|
Size: 3021
Comment:
|
Size: 4538
Comment:
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 1: | Line 1: |
| = How-To create the Pointing Plot = | = How To create the Pointing Plot = <<TableOfContents(5)>> |
| Line 3: | Line 4: |
| == Example Plot == {{attachment:plPoiFoc.png|width=70%}} |
|
| Line 6: | Line 11: |
| First off all you need the up-to-date version of the python script "plPoiFoc.py" that creates the plot. | First off all you need the up-to-date version of the python script "pointFoc.py" that creates the plot. |
| Line 15: | Line 20: |
| == Get the Pointing results from Projects == | == Get the Pointing results from Projects == |
| Line 19: | Line 24: |
| Please ask the computer group or me for the login details in case you don't know. | Please ask the computer group or me (CB) for the login details in case you don't know. |
| Line 21: | Line 26: |
| After logging in go to the search form and enter the time range you want | After logging in, go to the search form and enter the time range you want |
| Line 25: | Line 30: |
| Now you'll obtain a somewhat confusing list of scans and projects that have been observed during that time: |
|
| Line 28: | Line 31: |
| To get the pointing results and the data format the program reads you have to select one project like shown in the previous image (Project 266-11). Clicking on a project name leads you to this page: |
|
| Line 31: | Line 32: |
| Where you have to selcet the link '#All Scans' like indicated in the image. Now you'll get a list of all scans made in this project. |
{{attachment:searchForm2.png||width=70%}} |
| Line 34: | Line 34: |
| Note, that thus the scans may excced the time range you have selected at the beginning. Long projects like the flux-monitoring easily cover months, if not years. |
|
| Line 37: | Line 35: |
| On the top of the page you can adjust the number of scans shown by changing the number of Rows. This number should be always larger than the number of total scans to get all the scans. Then to get the information in the format the pyhton script accepts you have to change "Format" to CSV. This results in a pop up that is also displayed in the previous image. |
|
| Line 41: | Line 36: |
| == Creating the input File for the Python Script == The information in the format displayed in the pop up is what the python script expects. To get the script to work you'll need to create a text file that is called exactly: '''pointFocus.txt''' Into this text file you just need to copy the output displayed in the pop-up created by TAPAS. Important is that you '''don't''' copy the first three lines, i.e. the line staring with "Project" the empty line and the one starting with "Scan". It is also important that there are no empty lines in pointFocus.txt! == Creating the Plot== To create the plot, once you have the text file ready. You have to have '''plPotFoc.py''' and '''pointFocus.txt''' in the same directory and execute in the terminal: ''' python plPotFoc.py ''' This will print out the mean pointing and focus corrections on the screen. And create the pointing plot named: '''plPoiFoc.eps'''. == Plotting more than one project== To plot more than one project you just have to add the content of the CSV pop-up of the projects to '''pointFocus.txt''', every time without the first three lines and letting no line empty: |
Now you'll obtain a somewhat confusing list of scans and projects that have been observed during that time: |
| Line 73: | Line 43: |
| {{attachment:selectProject2.png||width=70%}} To get the pointing results and the data format the program reads you have to select one project like shown in the previous image (Project 266-11). Clicking on a project name leads you to this page: {{attachment:scan2.png||width=70%}} Here you have to select the link '#All Scans' like indicated in the previous image. Now you'll get a list of all scans made in this project: {{attachment:List2.png||width=70%}} Note, that the scans may exceed the time range you have selected at the beginning, since this page displays all scans of a project. Long projects like the flux-monitoring easily cover months. On the top of the page you can adjust the number of scans shown by changing the number of Rows. This number should always be larger than the number of total scans to assure that all scans are displayed. Finally, to get the information in the format the python script accepts you have to change "Format" to CSV. This results in the pop-up that is also displayed in the previous image. == Creating the input File for the Python Script == The information and format displayed in the CSV pop-up is what the python script expects. To get the script to work you'll need to create a text file that is called exactly: '''pointFocus.txt''' Please then copy the output displayed in the pop-up created by TAPAS into this text file. == Creating the Plot == '''Important''' for the script to work is that the PC that you are using has numpy and matplotlib, two python packages, installed. At the moment, without gra-lx4 available, only '''gra-lx18''', my machine, has the right preferences, as far as I know. '''Update by AS:''' Now the script can also be used on the '''mrt-aod''' and '''mrt-lx3''' computers. To create the plot, once you have the text file ready, you have thus to log into gra-lx18 and having '''pointFoc.py''' and '''pointFocus.txt''' in the same directory you need to execute in a terminal: ''' python pointFoc.py ''' This will print out the mean pointing and focus corrections on the screen and create the pointing plot named: '''plPoiFoc.eps''' in the same folder. == Plotting more than one project == To plot more than one project you just have to add the content of the CSV pop-up of the all projects to the same '''pointFocus.txt'''. Just append the new scans to the bottom of the already existing text file. The script orders the scans in time on its own. Don't worry about empty lines or lines that have not the right format since they are filtered out by the script. == In case of Problems == If you get error messages and/or fail to create the plot, please check if you have the latest code. If so you may send me (CB, buchbend AT iram.es) the pointFocus.txt you use and the name of the computer on which you tried to execute the script. Then I can check what is going wrong. == Future Ideas == We plan to develop a script that directly accesses TAPAS database and thus is much easier to handle. The plan is to make it work like the old MOPSIC commands, giving only the dates between one wants to examine the plot. Later this may become even an online tool within tapas. Until then please stick with this somewhat cumbersome way of creating the plot. However, once used to it, it does not take more than 5 minutes. |
How To create the Pointing Plot
Contents
Example Plot
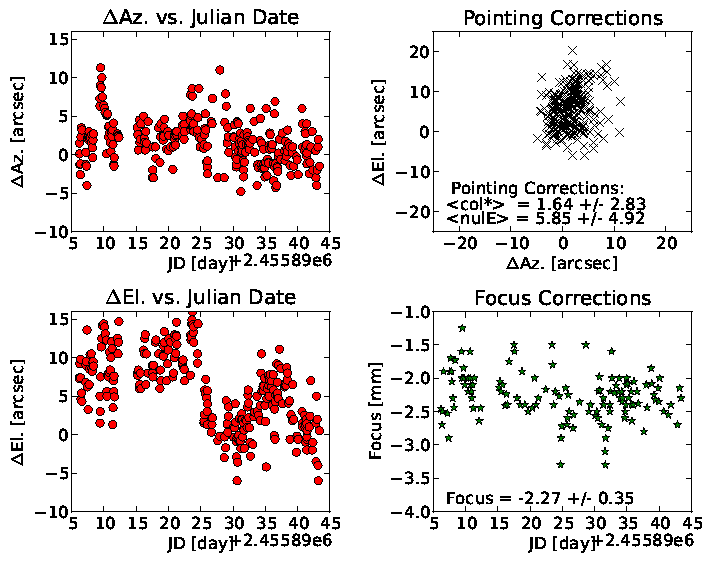
Download the Python Script
First off all you need the up-to-date version of the python script "pointFoc.py" that creates the plot. This can be downloaded here:
http://bazaar.launchpad.net/~ascurion/+junk/plPoiFoc/files
If you want you can contribute to the code via launchpad or send me changes you made so that I can include them. Any suggestions and improvements are welcome.
Get the Pointing results from Projects
Enter the Tapas webpage: https://mrt-lx3.iram.es/tapas/ . You'll have to log in with staff permissions to see the information of all projects. Please ask the computer group or me (CB) for the login details in case you don't know.
After logging in, go to the search form and enter the time range you want to display the pointing results for:
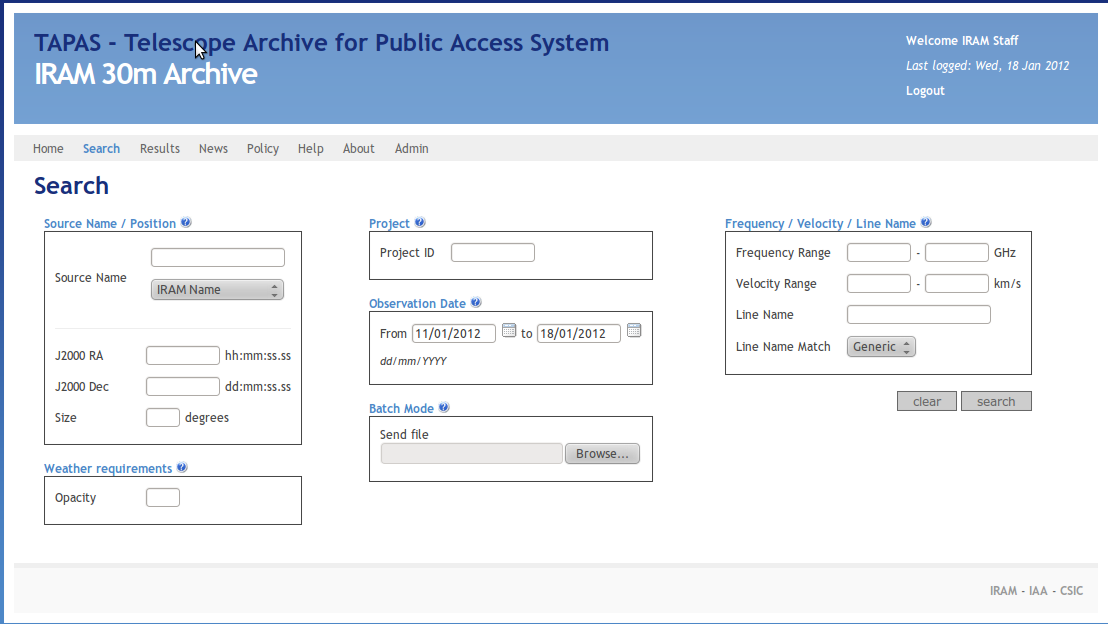
Now you'll obtain a somewhat confusing list of scans and projects that have been observed during that time:
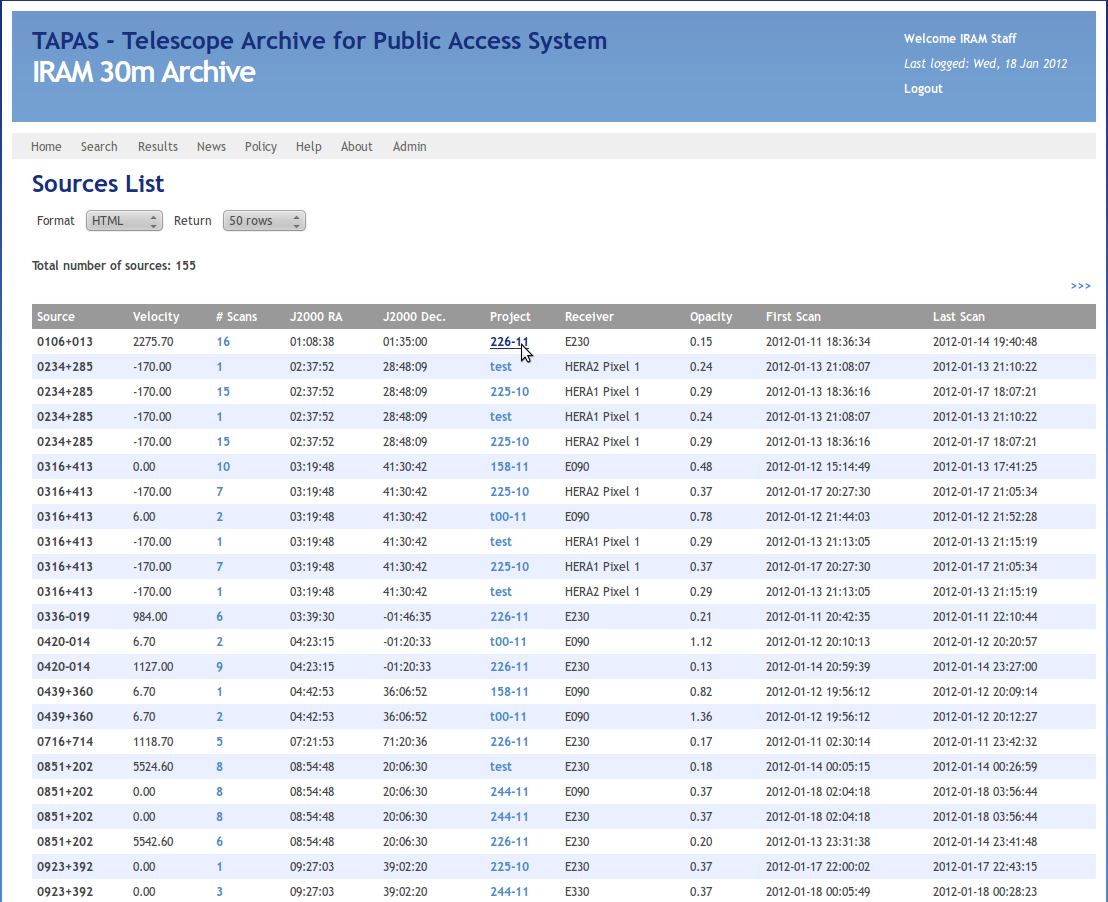
To get the pointing results and the data format the program reads you have to select one project like shown in the previous image (Project 266-11).
Clicking on a project name leads you to this page:
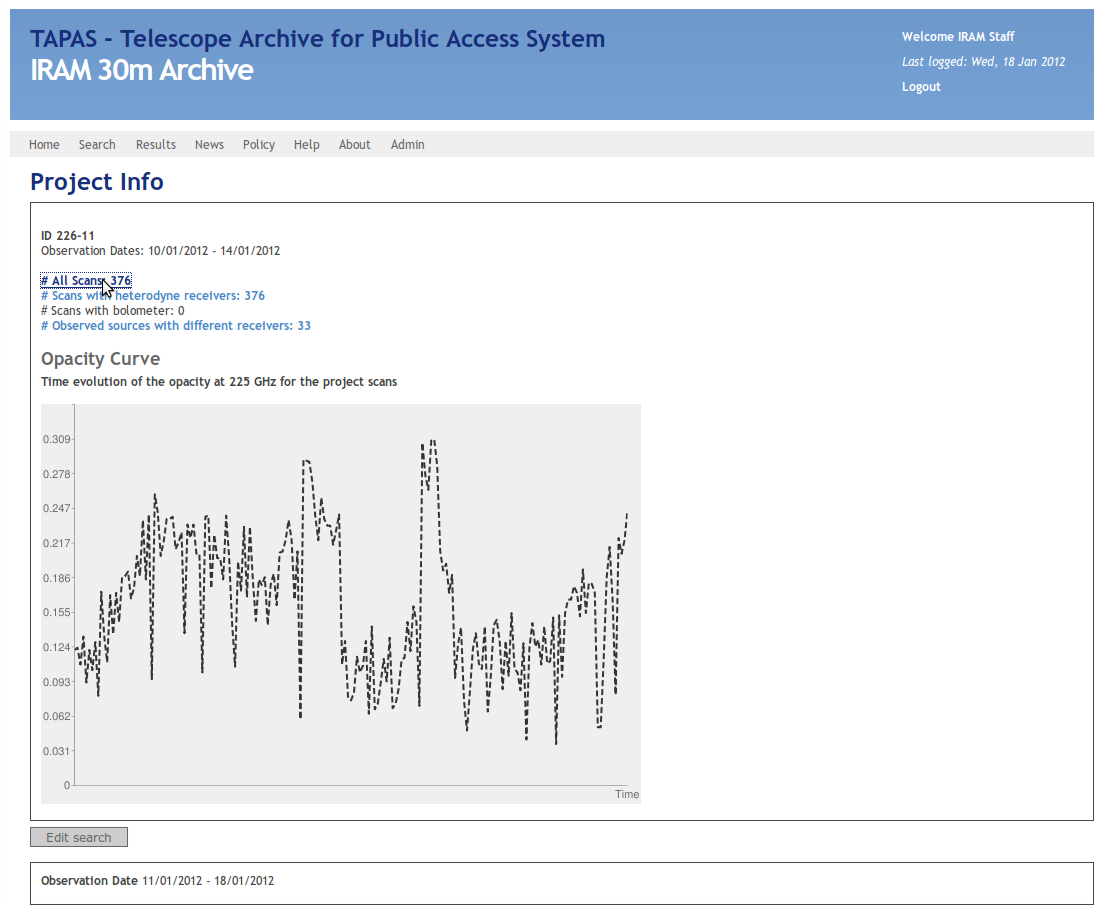
Here you have to select the link '#All Scans' like indicated in the previous image. Now you'll get a list of all scans made in this project:
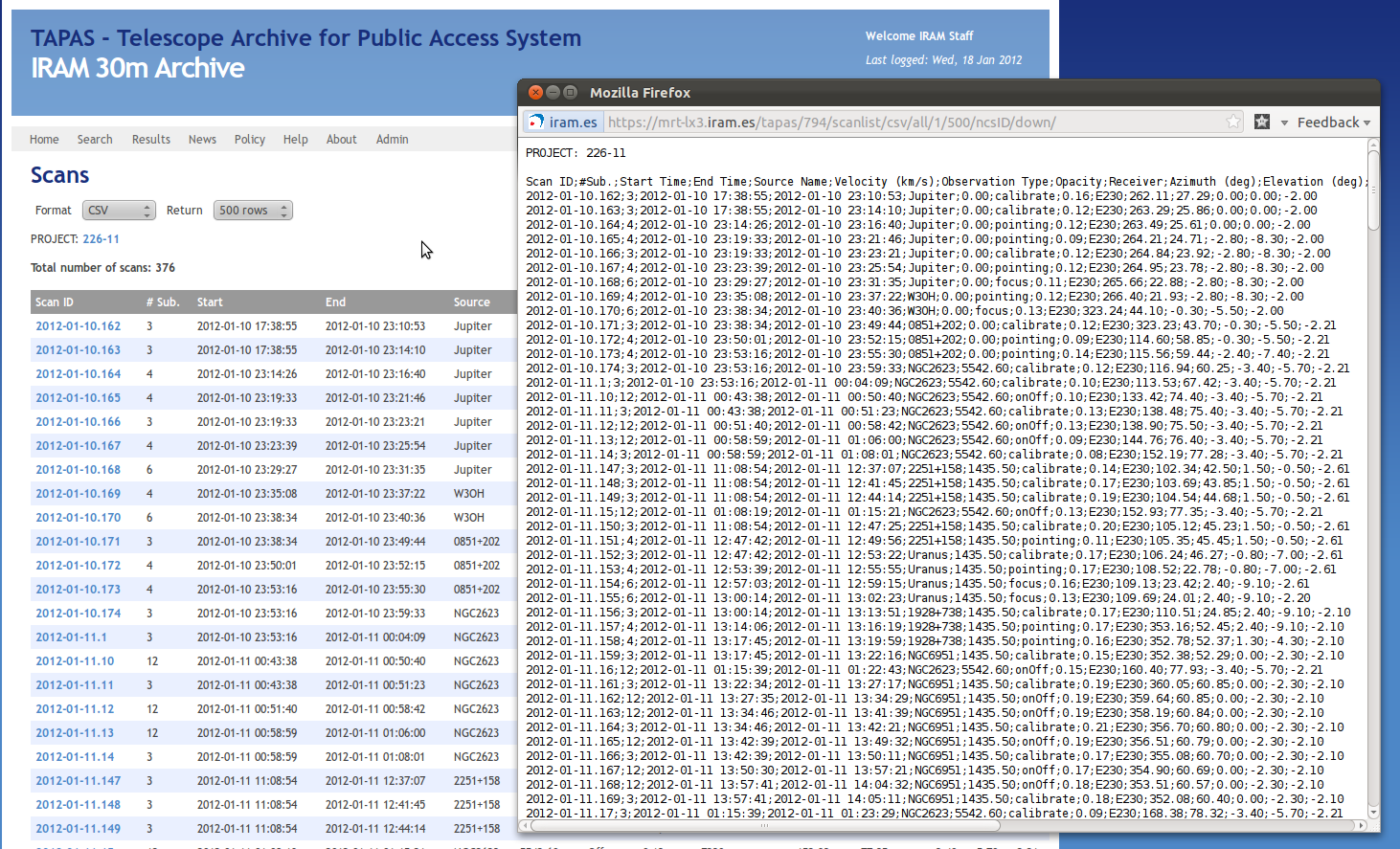
Note, that the scans may exceed the time range you have selected at the beginning, since this page displays all scans of a project. Long projects like the flux-monitoring easily cover months.
On the top of the page you can adjust the number of scans shown by changing the number of Rows. This number should always be larger than the number of total scans to assure that all scans are displayed. Finally, to get the information in the format the python script accepts you have to change "Format" to CSV. This results in the pop-up that is also displayed in the previous image.
Creating the input File for the Python Script
The information and format displayed in the CSV pop-up is what the python script expects. To get the script to work you'll need to create a text file that is called exactly:
pointFocus.txt
Please then copy the output displayed in the pop-up created by TAPAS into this text file.
Creating the Plot
Important for the script to work is that the PC that you are using has numpy and matplotlib, two python packages, installed. At the moment, without gra-lx4 available, only gra-lx18, my machine, has the right preferences, as far as I know.
Update by AS: Now the script can also be used on the mrt-aod and mrt-lx3 computers.
To create the plot, once you have the text file ready, you have thus to log into gra-lx18 and having pointFoc.py and pointFocus.txt in the same directory you need to execute in a terminal:
python pointFoc.py
This will print out the mean pointing and focus corrections on the screen and create the pointing plot named: plPoiFoc.eps in the same folder.
Plotting more than one project
To plot more than one project you just have to add the content of the CSV pop-up of the all projects to the same pointFocus.txt. Just append the new scans to the bottom of the already existing text file. The script orders the scans in time on its own. Don't worry about empty lines or lines that have not the right format since they are filtered out by the script.
In case of Problems
If you get error messages and/or fail to create the plot, please check if you have the latest code. If so you may send me (CB, buchbend AT iram.es) the pointFocus.txt you use and the name of the computer on which you tried to execute the script. Then I can check what is going wrong.
Future Ideas
We plan to develop a script that directly accesses TAPAS database and thus is much easier to handle. The plan is to make it work like the old MOPSIC commands, giving only the dates between one wants to examine the plot. Later this may become even an online tool within tapas. Until then please stick with this somewhat cumbersome way of creating the plot. However, once used to it, it does not take more than 5 minutes.
