|
Size: 18872
Comment:
|
← Revision 54 as of 2009-12-09 16:38:41 ⇥
Size: 18774
Comment:
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 7: | Line 7: |
| The MOPSIC executable can be downloaded with the data via the IRAM Data Base (see below) or as a stand-alone-system from the IRAM Grenoble GILDAS download area (http://www.iram.fr/IRAMFR/GILDAS/). | The MOPSIC software can be downloaded from the official GILDAS webpage [[http://www.iram.fr/IRAMFR/GILDAS/]]. A somewhat outdated but still very useful cookbook of 1998 is the [[http://www.iram.es/IRAMES/otherDocuments/manuals/Datared/pockcoo.ps|Pocket Cookbook for the MOPSI Software by R.Zylka]] |
| Line 14: | Line 16: |
| === Tar-file structure of a full download (dafs, scripts, software & data) === Data can be dowloaded via GetData from the data base. You will be asked whether to include MOPSIC Scripts, MOPSIC DAFs, MOPSIC Software. If you have downloaded MOPSIC together with the DAFs (Data Associated Files) and the MOPSIC scripts used at the telescope, unpacking the tar file will generate six directories: (./) dafs (./) data (./) logs (./) mopsic (./) scripts (./) work |
=== Tar-file structure of a full download (dafs & data) === Data can be dowloaded via GetData from the data base. You will be asked whether to include the MOPSIC DAFs. If you have downloaded the data with the DAFs (Data Associated Files) unpacking the tar file will generate six directories: (./) dafs (./) data (./)info (./) logs (./) rcps (./) work |
| Line 25: | Line 27: |
The '''scripts''' directory contains all up-to-date MOPSIC scripts used in the database pipeline reduction (for details see below). |
|
| Line 188: | Line 188: |
| {{attachment:pointing-mopsic.png}} | {{attachment:mopsic-display03.png}} |
| Line 191: | Line 191: |
{{attachment:mopsic-display02.png}} |
MOPSIC is a software which has been developed and is constantly upgraded by Robert Zylka (IRAM, France). It was created to analyze bolometer data but can be used for much more versatile tasks.
This cookbook is to help for the MAMBO2 reduction with the pool data. It is largely taken from the READMEs provided by the pool system when downloading the project data and written by R. Zylka. It may help you as well to setup your observations.
The MOPSIC software can be downloaded from the official GILDAS webpage http://www.iram.fr/IRAMFR/GILDAS/.
A somewhat outdated but still very useful cookbook of 1998 is the Pocket Cookbook for the MOPSI Software by R.Zylka
Back to Pool page: PoolObserving
General Informations
Tar-file structure of a full download (dafs & data)
Data can be dowloaded via GetData from the data base. You will be asked whether to include the MOPSIC DAFs. If you have downloaded the data with the DAFs (Data Associated Files) unpacking the tar file will generate six directories:
![]() dafs
dafs ![]() data (./)info
data (./)info ![]() logs
logs ![]() rcps
rcps ![]() work
work
The dafs directory contains all calibration files and links needed by MOPSIC to calibrate your data. Zenith opacities, receiver channel (i.e. bolometer) parameters, bad receiver channels, flat-field for sky noise filtering and calibration factors are stored in MRT_MAMBO.TAUS, MRT_MAMBO_MB.RCP, MRT_MAMBO_PF.RCP, MRT_MAMBO.DRC, MRT_MAMBO.CSFF and MRT_MAMBO.CAL, ordered by the Julian Date or UT date.
The data directory contains all project observations (scans) as well as pointing, focus, calibration and skydips scans done within one hour from the target observations.
The logs directory contains a project summary listing all target and calibration scans. In mopsic you will find the MOPSIC executable (since June 15 2005 the f95 version).
The work directory contains a listing (mydata.LIST) of all scans in the data directory. Such list are use by MOPSIC to find and list data for reduction. Furthermore the directory contains links to the MOPSIC scripts in the script directory. If you are using our standard installation of MOPSIC you should use this directory to reduce the data.
MOPSIC installation
System requirements:
- standard Linux system
- March 2008 GILDAS ifort version installation required
Installation: If you have downloaded together with the MOPSIC software the DAFs-files MOPSIC will know all parameters related to the bolometer array calibration such as the gain parameters, array geometry and the counts per Jansky conversion as well as the zenith optical depths for all data obtained in pooled observations since summer 2001 (the optical depths since Feb 1999). Older DAFs are available under request. We therefore recommend to download the DAFs and to update them regularly. In order to use these parameters on your local MOPSIC installation you have to define the system variable MOPSIC_DAFS by adding the following line to your .cshrc or .tcshrc system file:
setenv MOPSIC_DAFS /YOUR/DOWNLOAD/INSTALLATION/PATH/dafs
or to the.bashrc file:''
export MOPSIC_DAFS=/YOUR/DOWNLOAD/INSTALLATION/PATH/dafs
To run MOPSIC from all directories on your system you furthermore need to add to the same file the path where the MOPSIC executable is located:
in .cshrc or .tcshrc:
set path path=($path /YOUR/DOWNLOAD/INSTALLATION/PATH/mopsic)
or in .bashrc:
PATH=$PATH:/YOUR/DOWNLOAD/INSTALLATION/PATH/mopsic export PATH
Of course it is not mandatory to keep MOPSIC and the DAFs in the directories created by the tar. You may put them wherever you want.
MOPSIC pipeline scripts
defs_general.mopsic script: The database pipeline reduction scripts are developed to be easy be used on your local PC. To do so, you have to edit the script. Simply change the logicalforiramdbto false if it was set to true:
let foriramdb .false. ! or let foriramdb no
This will set the input data directory and the default input list to the directory structure generated by the tar file and all DAFs to defaults, i.e. to the entries defined by MOPSIC_DAFS.
The main routines used for the data reduction are ooCSF.mopsic and mapCSF.mopsic
For some special purposes also special scripts are offered. They are obtained by changing some default variables set in map_defs or oo_defs correspondingly. Follow the suggestions described there.
Mapping
mapCSF.mopsic : (gridded phase difference - correlated signal filter general script formap reduction with sky noise subtraction
mapNcsf.mopsic : (gridded phase difference - no correlated signal filter) map reduction without sky noise subtraction
mapCSFrmx.mopsic : special scripts to reduce maps of relatively compact sources, <200" in diameter; it is not suitable for more extended and structureless sources but should give better sky noise subtraction for relatively compact sources.
mapCSF0.mopsic : map reduction with sky noise subtraction for sources which are weak (for CSF) and more extended than 0.5*beam_separation; weak for CSF means with peak fluxes < ~5*rms in the final data of one bolometer (i.e. ~5*30 mJy/beam)
mapCSFrmn_saa.mopsic: (gridded phase difference - correlated signal filter - shift and add) source structure is assumed to be smaller than 0.5*beam_sep.
OnOff
ooCSF.mopsic : (onoff - correlated signal filter) reduces onoffs with sky noise subtraction
ooNcsf.mopsic: (onoff - no correlated signal filter) reduces onoffs without sky noise subtraction
Both on-off scripts write the final results to 'source'.OOxx files. For solar system sources ("body") the corresponding scripts oo_ncsf_body and oo_csf_body are also within the tar. Note that the only one change between the "body" and the standard on-off scripts is the additional argument body within the oo-command.
MOPSIC uses a special method to calculate the chopped signal. This gives much better results than the straight difference of the signals obtained at both chopper positions. The method is described in the Dec. 2005 IRAM Newsletter. For some special purposes also special scripts are offered. They are obtained simply by changing some default variables set in map_defs and/or oo_defs correspondingly.
There is also a script ooCSF_multsou.mopsic whic allows to reduce on-offs of many sources. The user has to define the number of sources and the lists with the files. Those can be created from a general lis using the commad find objects write.
Misc
In addition there are also scripts to reduce pointings (p_nshav.mopsic - without sky noise subtraction, p_shav.mopsic - with very simple sky noise subtraction), skydips (skydips.mopsic) and to calculate the RCPs (Receiver Channel Parameters) from calibration maps. T
he scripts in the subdir specials are special versions of the general pipeline scripts. They use some other settings than defined in map_defs (see the corresponding map_defs_*). They are therefore optimized for some special purposes (e.g. subtracting a source distribution before reduction, applying base level corrections subscan by subscan, using polygons to define the base range, ...).
In the subdir extras you will find some scripts useful for checking (e.g. chk_rc) or just displaying the data. The snr... scripts demonstrate how to calculate a signa-to-noise ratio map.
In the subdir simple you will find a script map.mopsic which can be used to reduce maps of simple sources, e.g. compact or extended. It should not be used if the source shows both compact and extended components !
The script map_negres.mopsic allows to create a map with just the negative residuals of the source due to the shift-and-add restoring. Subtract the map produced by this script from the saa map. For further informations read the suggestions in README_oo, README_maps, README_glossary, oo_defs.mopsic, map_defs.mopsic. Furthermore, some special suggestions are given in some dedicated scripts (e.g. in mask_subsc_nr.mopsic, oo_oo.mopsic).
First Steps
To start MOPSIC type mopsic. Macros are called with @scriptname WEAKSOU (no extension needed). The argument WEAKSOU may have values no or yes and allows to optimise the data processing for weak sources, i.e. sources with peak flux densities below ~50mJy/beam in case of on-offs or ~200mJy/beam in case of maps. If you are using the default download structure you can reduce your data straight forward by calling your preferred script, e.g:
@mapCSF WEAKSOU ! WEAKSOU may be yes or no
or
@ooCSF WEAKSOU ! WEAKSOU may be yes or no
MOPSIC will read all observations from the mydata.LIST, select the corresponding type of observations (i.e. maps or on-offs) and display a list of these selected observations. Within the reduction scripts you have different possibilities to select the data you may want to process:
select name ! select all observations of a specific source (combined result) select name* ! select similar source names (e.g. to create a mosaic of maps or on-offs done on different parts of a source) select no name[*] ! excludes observations from the LIST with defined source names (do not use the exclude command, it is not finished)
Alternatively you can delete some observations from the list, e.g.:
delete file iram30m-abba-20060118s85 ! remove this observation from the list delete scan 20060118s85 !remove the observation with the scan number 85 observed on jan 18 2006 from the list.
To have a help online just type:
? select or ? delete !lists all possibilities
The command find objects [> #, write] might also help.
Typing c will continue the reduction. The final result will be written to a FITS file in case of mapping. The default output name is scriptname.fits. Note that MOPSIC will not overwrite any results obtained in a previous reduction, except for GDF-files. If you use the same script for a different source make sure you move the FITS file to a different name before starting the script again or - the better solution - create dedicated scripts for each source and change the default fits name. IF YOU DO NOT CARE OVERWRITING the prevoius results, use the GDF files as output.
To view the results on a graphic device, open the device (before or after you execute the script:
device image white (or black) lut idl4 ! Choose color table
for maps:
plot [options] , e.g. plo scaling linear -30 100
for onoffs:
plot oo [options].
There are also some pl... scripts, which might help checking the data quality.
Since MOPSIC is under developement and still not fully integrated within the GILDAS software, no detailed manual is available so far. A help on individual commands, however, is available by typing: ? command or help command for GILDAS commands.
Almost all commands used for advanced data reduction can be found in the MOPSIC scripts. MOPSIC, however, offers a much broader range of applications including advanced planning functions for mapping and onoff observations, post-reduction data analysis and processing and even reduction of non-bolometer data (optical, IR, spectroscopy). If you have questions how to obtain the best results from your data do not hesitate to contact Robert Zylka (zylka@iram.fr ).
Mopsic Displays
Figure 1. ONOFF Online Display
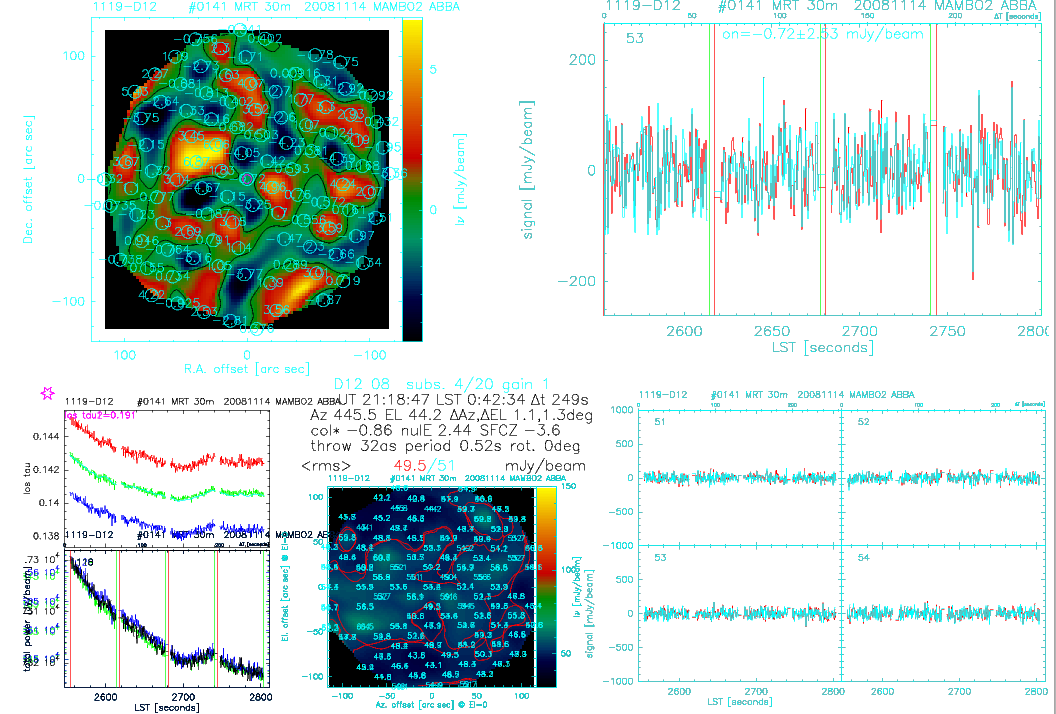
- Upper left: on-off RA/DEC map of all pixels. The RA/DEC 0/0 denotes the reference pixel for pointing.
- Upper right: center pixel (53): before/after skynoise reduction, i.e. correlated signal filter (CSF, red/green)
- Lower left:
- A star denotes the result of the last sky dips, i.e. the sky emissivity at the source elevation CHECK
- Output of 3 DC pixels to check for changes in the sky emissivity
- upper: line of sight (los) opacity (showing elevation effect and others)
- lower: total power
- Lower center:
- noise in the data before/after the CSF (the plot before is/will be overplotted with the noise after the CSF, in this particular case the rms in the data after CSF is visible, the numbers above the plot 49.5/51mJy/beam give the rms values of the high frequency noise component)
- gain 1 (in this particular case)
- col*/nule/sfcz, i.e. pointing and focus offsets
- throw 32", period 520msec, rotator angle 0
- Lower right: time series of 4 pixels (51, 52, 53, 54), before and after sky noise subtraction
Figure 2. Pointing Online Display
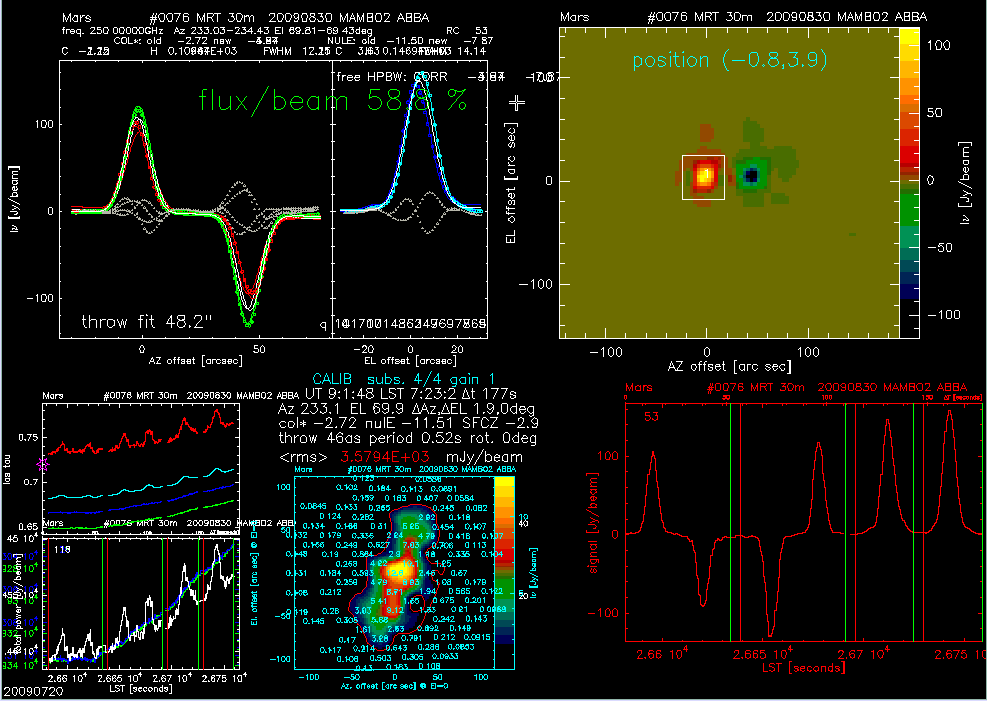
Figure 3. Skydip Online Display
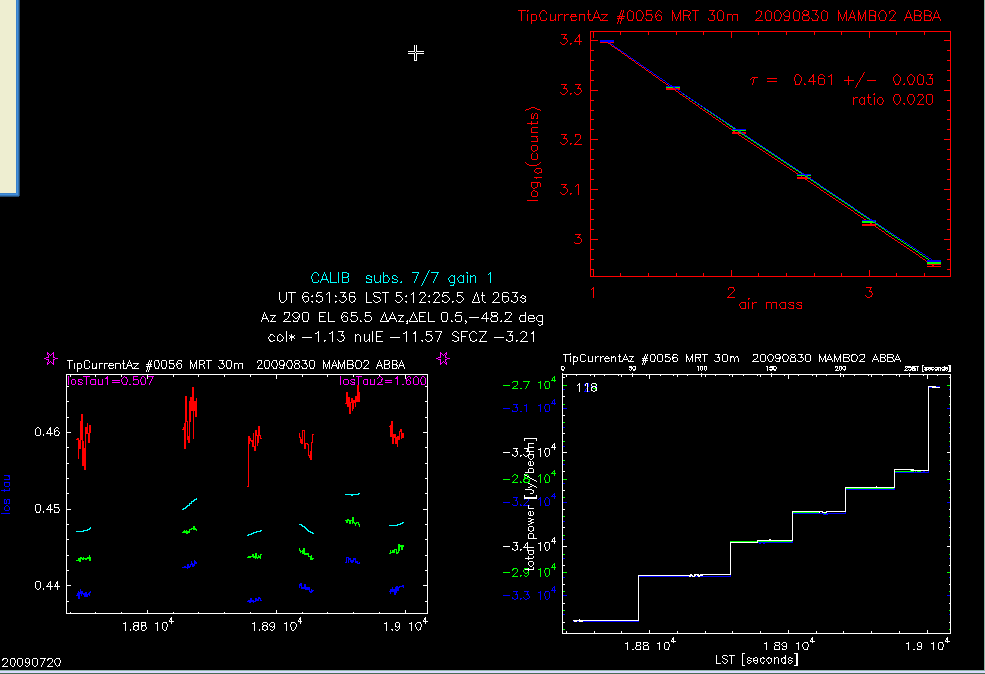
Figure 4. ONOFF data reduction: Results
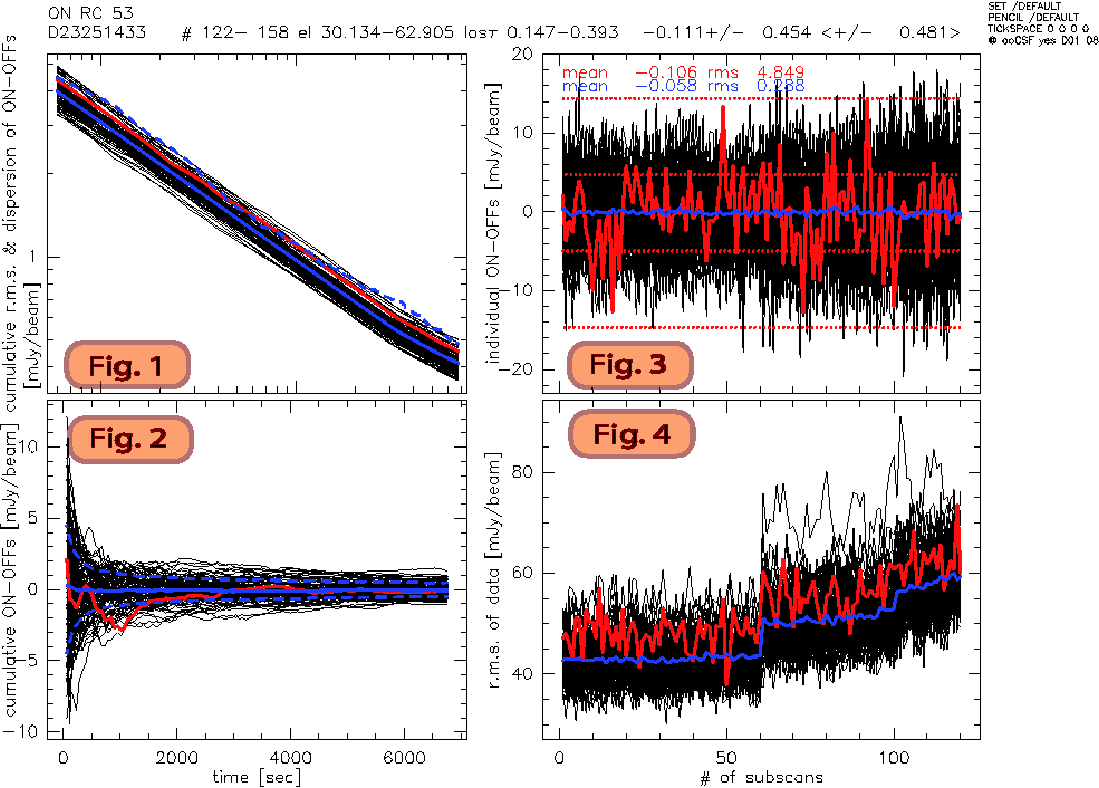
subFig 1: Noise vs Time
subFig 2: Is the average of Fig 3 vs time
subFig 3: Red represents the central pixel (20 or 53) per # of subscans.
subFig 4: Total power on the pixels (red is central pixel) after correlated noise removal.
Appendix
Notes for maps
Look at NoteMopsicMaps for some detailed explanation on the settings of the reduction.
Notes for OnOff
Look at NoteMopsicOnoff for some detailed explanation on the settings of the reduction
Mopsic Glossary
Some useful commands of MOPSIC:
inDir FILE defines the directory for input data
outDir FILE defines the directory for output data
inList FILE loads the observational parameters from FILE.LIST
base in time btorder performs polynomial fit of order btorder to the time sequences, i.e. to signal = f(time)
base in subscans blorder performs polynomial fit of order blorder to signals as function of the scanning coordinate, subscan by subscan (i.e. row by row in a map)
calibrate FILE reads the calibration data cal_factor(JD) from the file FILE; the units might be counts/Jy or Jy/counts
calibrate default reads the calibration data defined in the DAFs, i.e. the calibration data base
calibrate dividing signal[Jy/beam]=signal[counts/beam]/cal_factor[counts/Jy] as function of the Julian date JD; the calibration data must be read in before
calibrate multiply signal[Jy/beam]=signal[counts/beam]*cal_factor[Jy/counts] as function of the Julian date JD; the calibration data must be read in before
calibrate back signal[counts/beam]=signal[Jy/beam]*cal_factor[counts/Jy] or signal[counts/beam]=signal[Jy/beam]/cal_factor[Jy/counts] as function of the Julian date JD; the calibration must have been applied before
convert [options] converts either (Az,EL) to (RA,DEC) coordinates or (RA,DEC) to (l,b)
correct extinction corrects for the atmospherical extinction, the zenith optical depth values must be loaded before (see the tau-command)
correct ge corrects for the gain-elevation dependance
delete dc deletes the data of the "total power" bolometers
delete rc default deletes the data of the bolometers which are defined in the DAFs, i.e. the calibration data base
finish finishes a subprogram, e.g. CONVERT, CSF, COMBINE, AE_OBS, REPROJECT, ...
focus reduces the focus
grid xinc grids the data in the scanning direction only, i.e. no 2dim or 3dim gridding
plot [something] plots either the map in memory or sometihig else; the plotting device must be defined before (devide command)
pointing reduces the pointing
read ... /phases reads the data of both wobbler positions (the "phases") as independent time sequences
read ... /pha dif reads the data of both wobbler positions and calculates the sequential differences as follows signal(i) = phase_left(i)-phase_right(i)
rms [options] calculates the rms in the data
run executes a subprogram, e.g. CONVERT, CSF, COMBINE, REPROJECT, ...
scan # [/name NAME] reads raw data of scan # The option /name must be defined at least once or the scan name NAME must be set with set scan_name. The scan name NAME must include a * at the position of the scan # and the extension defining the format, e.g. iram30m-abba-20060120s*-imb.fits
show rcp plots and prints the Receiver Channel Parameters
select something selects the files from the input list (loaded with the command in_list) according to the defined selection criteria
set scanName NAME sets the name of files for the command scan. The scan name NAME must include a * at the position of the scan # and the extension defining the format, e.g. iram30m-abba-20060120s*-imb.fits
set scanDate DATE sets date in the file name used with the scan command; the format of DATE is YYYMMDD, e.g. 20060121
store [something] stores either the current map in the backup buffer or something, e.g. the weighting
tau default timeInterval reads the zenith optical depth values from the file defined in the DAFs, i.e. the calibration data base
write fileName [/format formatName] writes the data to the file fileName in format determined by the extension of fileName, e.g. gdf, fits ... [in format specified by the option /format formatName]
write +addName [/format formatName] writes the data to the file with name = inputDataFileName+addName
CSF Correlated Signal Filter; as the correlated signal should be mostly the sky noise, it filters out the sky noise, the csf-command calls the sub-program CSF
DBF Double Beam Filter (EKH, 1979)
DBR Double Beam Restoration (i.e. deconvolution, EKH, 1979)
> # [rms] clips the data above # [*rms]
< # [rms] clips the data below # [*rms]
